

 http://www.clearlight.com/~morph/dna.htm (Internet on a CD, 07/1998)
http://www.clearlight.com/~morph/dna.htm (Internet on a CD, 07/1998)See the Easier to Understand, Everyone's Guide to DNA Computers
Copyright 1996, Yali Friedman
All Rights Reserved
morph@clearlight.com
February 5th, 1996
On November 11 1994, a paper in Science[1] described the "Molecular Computation of Solutions of Combinatorial Problems". This was the first ever implementation of a DNA-based computer. Since then, many advancements have been proposed to refine the protocol for programming a DNA computer to reduce the complexity of the operations and eliminate errors. [2,3] Despite their respective complexities, biological and mathematical operations have some similarities:
For the same reasons that DNA was presumably selected for living organisms as a genetic material, its stability and predictability in reactions, DNA strings can also be used to encode information for mathematical systems.
Figure 1 shows a diagram of the Hamiltonian Path problem. The objective is to find a path from start to end going through all the points only once. This problem is difficult for conventional computers to solve because it is a "non-deterministic polynomial time problem" (NP). NP problems are intractable with deterministic (conventional/serial) computers, but can be solved using non-deterministic (massively parallel) computers. A DNA computer is a type of non-deterministic computer. The Hamiltonian Path problem was chosen because it is known as "NP-complete"; every NP problem can be reduced to a Hamiltonian Path problem.[4]
The following algorithm solves the Hamiltonian Path problem:
The key to solving the problem was using DNA to perform the five steps in the above algorithm. The following operations can be performed with DNA:
The above operations can be used to "program" a DNA computer.
To implement step 1 of the algorithm, Adleman created a 20-mer sequence of DNA for each city A through G. For each path i>j, an oligonucleotide was created that was the 3' 10-mer of i and 5' 10-mer of j (see figure 2). A>B contained all of A and the 5' 10-mer of B, and E G contained the 3' 10-mer of E and all of G to eliminate any possible sticky ends. For each city i, a Watson-Crick complementary oligonucleotide was synthesized and designated ~i.
For each city except A and G, and for each path, 50 pmol of and 50 pmol of i>j, respectively, were mixed together in a single ligation reaction. The oligonucleotides served as splints to bring paths between common cities to associate for ligation. The ligation reaction therefore resulted in the formation of DNA molecules encoding random paths through the graph.

Step 2 was implemented by using A and ~G as primers for PCR to amplify all "paths" from starting with A and ending with G.
For step 3, the product of step 2 was run on an agarose gel, and the 140 bp band (corresponding to a dsDNA path entering exactly seven cities) was excised and soaked in double-distilled H2O to extract the DNA. This product was then PCR amplified and gel-purified several times to enhance its purity.
In order to complete step 4, the product of step 3 was affinity-purified with a biotin-avidin magnetic beads system. The dsDNA from step 3 was melted and the ssDNA product was incubated with ~A conjugated to magnetic beads. Only those ssDNA molecules that entered city A at least once were annealed to the bound ~B and retained. The process was successively repeated with ~C, ~D, ~E, and ~F.
Step 5 was performed by amplifying the product of step 4 with PCR and running it on a gel. Figure 3 shows the results of these procedures. In figure 3A, lane 1 is the result of the ligation reaction in step 1. The smear is consistent with the construction of molecules encoding random paths through the graph. Lanes 2-5 show the results of the PCR reaction in step 2. The densest bands correspond to the amplification of molecules encoding paths that began at A and ended at F.
Figure 3B shows graduated PCR of the final product of the computer.
Graduated PCR allows one to "print" the results of a computation, using A as the
right primer and ~B through ~G as the left primer lanes 1-7, respectively. The graduation
shows the progression from A>B>C>D>E>F>G. 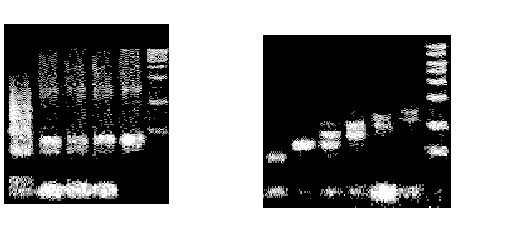
The above procedure took approximately one week to perform. Although this particular problem could be solved on a piece of paper in under an hour, when the number of cities is increased to 70, the problem becomes too complex for even a supercomputer. The fastest supercomputers can currently perform 1000 million instructions per second (MIPS); a single DNA molecule requires approximately 1000 seconds to perform an instruction (.001 MIPS). Obviously of you want to perform one calculation at a time (serial logic), DNA computers are not a viable option. However, if one wanted to perform many calculations simultaneously (parallel logic), a computer such as the one described above can easily perform 10^14 MIPS. DNA computers also require less energy and space. While existing supercomputers operate 10^9 operations per Joule, a DNA computer could perform 2 x 10^19 operations per Joule (10^10 times more efficient). Data can be stored on DNA at a density of approximately 1 bit per cubic nm, while existing storage media require 10^12 cubic nm to store 1 bit.[1]
Since Adleman's original experiment, several methods to reduce error and improve efficiency have been developed.[2,3,5] The problems with implementing a DNA computer can be separated into two types:
The Restricted model of DNA computing solves several physical problems with the Unrestricted model. The Restricted model simplifies the physical obstructions in exchange for some additional logical considerations. The purpose of this restructuring is to simplify biochemical operations and reduce the errors due to physical obstructions.
Despite these restrictions, this model can still solve NP-complete problems such as the 3-colourability problem, which decides if a map can be coloured with three colours in such a way that no two adjacent territories have the same colour.
Certain assumptions must be made about the oligonucleotides used in the manipulations:
Error control is achieved mainly through logical operations, such as running all DNA samples showing positive results a second time to reduce false positives. Some molecular proposals, such as using DNA with a peptide backbone for stability, have also been recommended.
Recently, Bartel and Szostak[5] used the methods of combinatorial chemistry to make a pseudo-enzyme. The goal of their experiment was to find a molecule of RNA which would ligate two substrate molecules of RNA. They used a pool of approximately 4^25 random sequences of RNA to ligate the two substrate molecules, and after isolating the product of the reaction, they were able to sequence the pseudo-enzyme.
The applications of combinatorial chemistry go far beyond this one example. In the above example, the pseudo-enzyme remained bound to the product, making it easy to isolate, but combinatorial chemistry can also be used when anchoring is not physically possible. Protocols have been proposed to find an RNA molecule that truly catalyzes (as an enzyme) the ligation of two DNA molecules. This sort of detection can also be used to isolate an endonuclease, or to find a drug which crosses a cell membrane and then binds a particular host membrane, and therefore cannot be anchored.[5]
DNA computing is less than two years old (November 11, 1994), and for this reason, it is too early for either great optimism of great pessimism. Early computers such as ENIAC filled entire rooms, and had to be programmed by punch cards. Since that time, computers have since become much smaller and easier to use. It is possible that DNA computers will become more common for solving very complex problems, and just as PCR and DNA sequencing were once manual tasks, DNA computers may also become automated.
In addition to the direct benefits of using DNA computers for performing complex computations, some of the operations of DNA computers already have[5], and perceivably more will be used in molecular and biochemical research.